So... what's Digital Pathology?
Adrien Foucart, PhD in biomedical engineering.
This website is guaranteed 100% human-written, ad-free and tracker-free. Follow updates using the RSS Feed or by following me on Mastodon
Adrien Foucart, PhD in biomedical engineering.
This website is guaranteed 100% human-written, ad-free and tracker-free. Follow updates using the RSS Feed or by following me on Mastodon
Let's start from the beginning. When I started my thesis, the topic we settled on was "Deep Learning in Digital Pathology". It's vague - but that was kind of the point. Deep Learning and Digital Pathology were both recent trends at the time, so trying to look at what could be done with it in general seemed like a good idea.
We have two parts in "Deep Learning in Digital Pathology". The first one, Deep Learning, is where I have spent most of my thesis. That's the part that concerns me as a biomedical engineer specialized in image analysis, and where I can contribute the most. The second part, however, is just as important. It's the application, what we want to use Deep Learning for: Digital Pathology. I am certainly not an expert in Digital Pathology - and even less of an expert in not-digital histopathology - but understanding what the methods I'm going to develop may be used for does seem like a good idea, so let's briefly get into it.
The goal of histopathology is to examine human tissue (usually taken from a tumour or some other possibly diseased area) under a microscope, to formulate a diagnosis or to get a better understanding of a disease. The process, in short, is as follows:
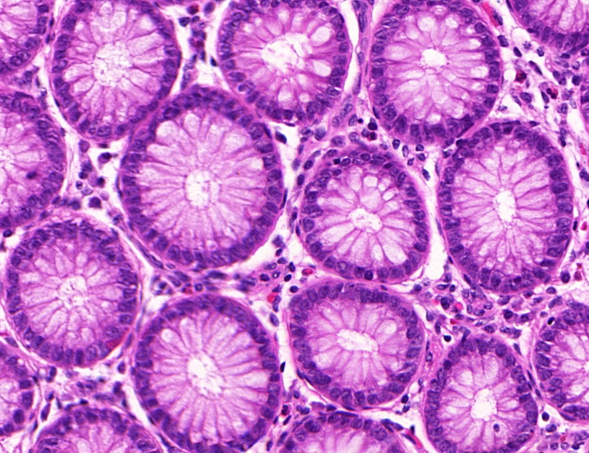
Why do we have to stain the tissue? Because cells are mostly water, water tends to be transparent [citation needed], and transparent things are hard to look at with visible light. Fortunately, some chemical pigments have properties which are very useful for pathologists. For instance, in the late 19th and early 20th century, it was discovered that we could use hematoxylin to stain the nuclei of cells in blue, and eosin to stain the cytoplasm in pink [1].
This produces "Hematoxylin & Eosin" - H&E - images like this one below, where the structure of the tissue is easy to analyse for the pathologist:
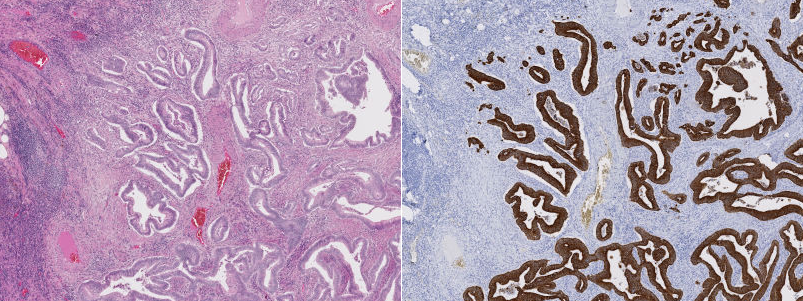
Slightly more recently, we realized that we could use the properties of antigens and antibodies to get some more specific staining. The idea is this: in our body, some cells - antibodies - are designed to specifically bind to some proteins - antigens - as a defence mechanism to produce an immune response. We can "hack" this process by binding a staining agent to an antigen and therefore "highlight", in the tissue, places where the related antibodies are present. This method is called immunohistochemistry, or IHC. For instance, in the image below, we have the same part of the tissue stained with H&E on one side, and with an IHC marker (anti-pan-cytokeratin, to be precise) on the other. The IHC marker highlights the cells which are part of a tumour, which is a rather useful information to have in histopathology.
So where does the "digital" part fit in all this?
The problem with the process above is that it requires the trained pathologist to be physically in front of the microscope, with the slide in it. There are a number of drawbacks to this. One is that it's hard to get a second opinion from a specialist from somewhere else. Another is that comparing a tissue to, for instance, another sample taken some months or years before requires finding the physical slide in the archives of the hospital.
How do we solve that? With digital scanners. Very expensive, very high resolution, very precise digital scanners. The entire slide can be scanned at multiple levels of magnification to produce Gigapixel images, which can be viewed on a computer. The pathologist can then access the image in a "virtual microscope".
And once you start to have the slides as digital objects, you open the door to many possibilities, from tools to easily annotate the slides (for instance, for teaching purposes, or simply to quickly document the reasoning behind a diagnosis) to automated analysis of certain aspects of the tissue. In particular, some quantitative analysis are very hard to do for a human expert in an objective manner (like evaluating "what percentage of the tissue shows this marker?"), yet relatively easy (or at least possible) to do for a well-designed algorithm.
Digital image acquisition are becoming commonplace, and associated image analysis solutions are viewed by most as the next critical step in automated histological analysis.
In the more than 10 years since Mulrane's paper, there has indeed been a wide range of image analysis applications in digital pathology. And, in more recent years, as in most image analysis applications, one type of strategy seems to have very quickly surpassed all others: Deep Learning.
I guess that's a teaser for the "next episode"?