A timeline of Deep Learning in Digital Pathology
Adrien Foucart, PhD in biomedical engineering.
This website is guaranteed 100% human-written, ad-free and tracker-free. Follow updates using the RSS Feed or by following me on Mastodon
Adrien Foucart, PhD in biomedical engineering.
This website is guaranteed 100% human-written, ad-free and tracker-free. Follow updates using the RSS Feed or by following me on Mastodon
In 2017, Geert Litjens and his colleagues from Radboud University published a comprehensive survey of deep learning methods in medical image analysis [1]. It's possible that they missed some early papers that could qualify as "Deep Learning": as I've written before, the boundaries of what is or isn't Deep Learning are unclear, and articles written before 2012 are unlikely to use that terminology. If such articles exist, however, they have been forgotten in the large pile of never-cited research that fails to be picked-up by Google Scholar, Scopus, or other large research databases. Litjens' survey therefore remains the reference on the matter.
So who was first to use "Deep Learning in Digital Pathology"? The turning point seems to come from the MICCAI 2013 conference in Nagoya, with two pioneering articles: Angel Cruz-Roa's automated skin cancer detection [2], and Dan Cireşan's mitosis detection in breast cancer [3].
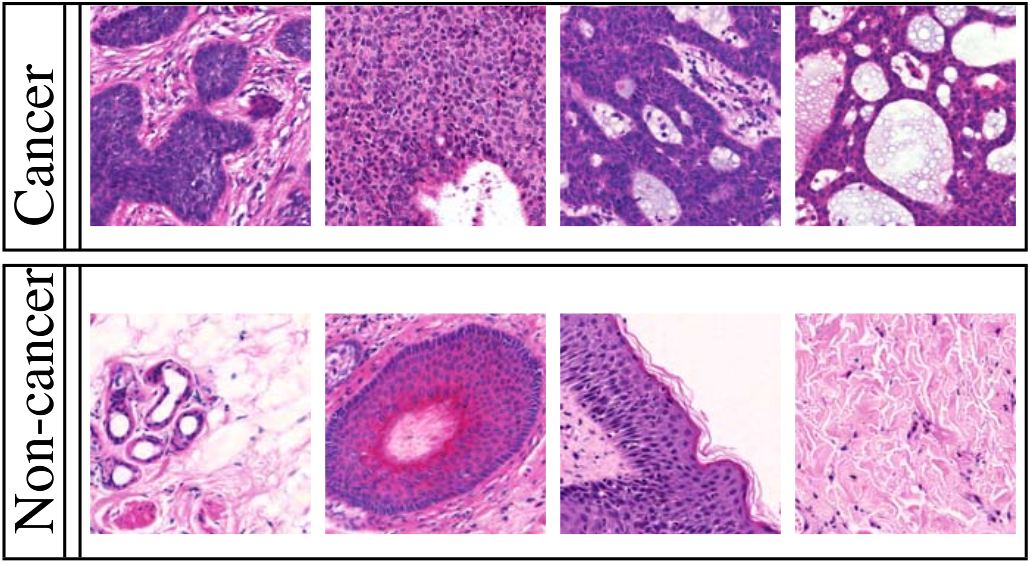
Cruz-Roa uses H&E stained images from skin tissue, to try to automatically determine if there is a malignant cancer in the sample. As illustrated in the figure below, the difference between cancer and non-cancer is based on morphological criteria which are very difficult to define.
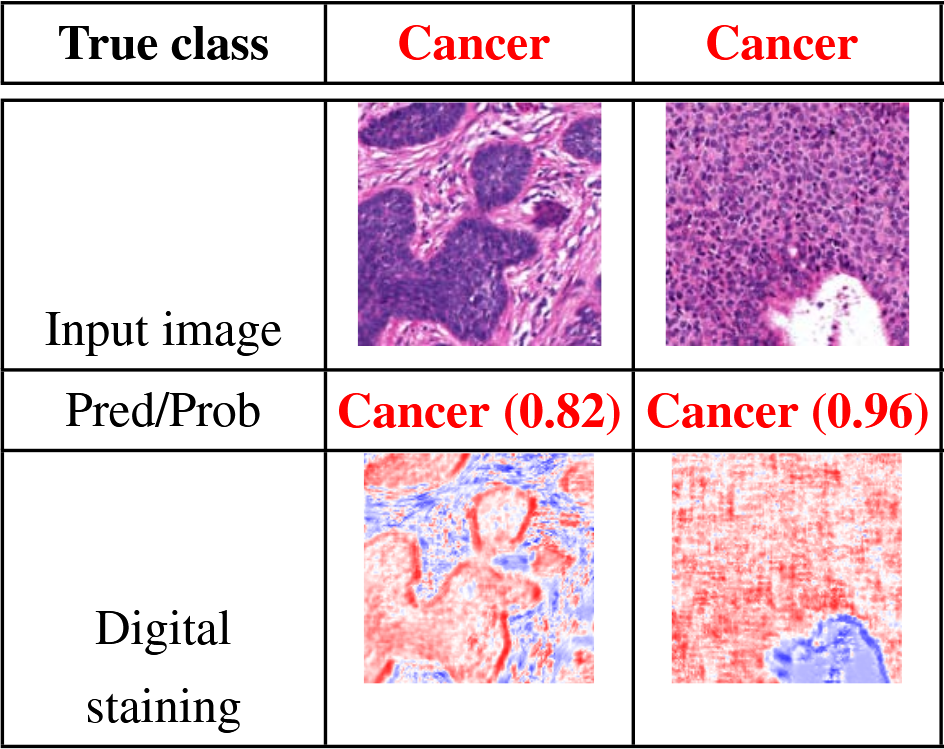
Their algorithm produces both a classification (cancer or not) at the image level, and what they call a "digital staining", which is basically a probability map of where the cancer regions are (see figure below). This is a very important feature for machine learning methods in biomedical imaging, related to the concept of interpretability. A machine learning algorithm which only produces a "diagnosis", but is unable to "explain" how this diagnosis came to be, cannot be trusted. I will most certainly come back to that idea later: the "reasoning" that machine learning algorithms (deep or not) make are sometimes more a reflexion of biases and artefacts in the data that was used to train it than of an understanding of the pathology. Having an output which includes an explanation of the diagnosis is therefore essential to control whether any "weird stuff" is happening.
Cireşan also uses H&E images, taken from breast cancer tissue. I'll let him introduce the problem:
Mitosis detection is very hard.
Mitosis - the process of cell multiplication - is a relatively rare event, meaning that, in the images which are available to train the algorithm, only a very small fraction will be part of a nuclei, and an even smaller fraction will be part of a nuclei going through mitosis. In addition to being rare, the appearance of the cell nucleus will be very different depending of which stage of the mitosis the cell is currently experiencing. To get an idea of how difficult the task is, we can just look at these examples from the article (click on the image for a larger version):
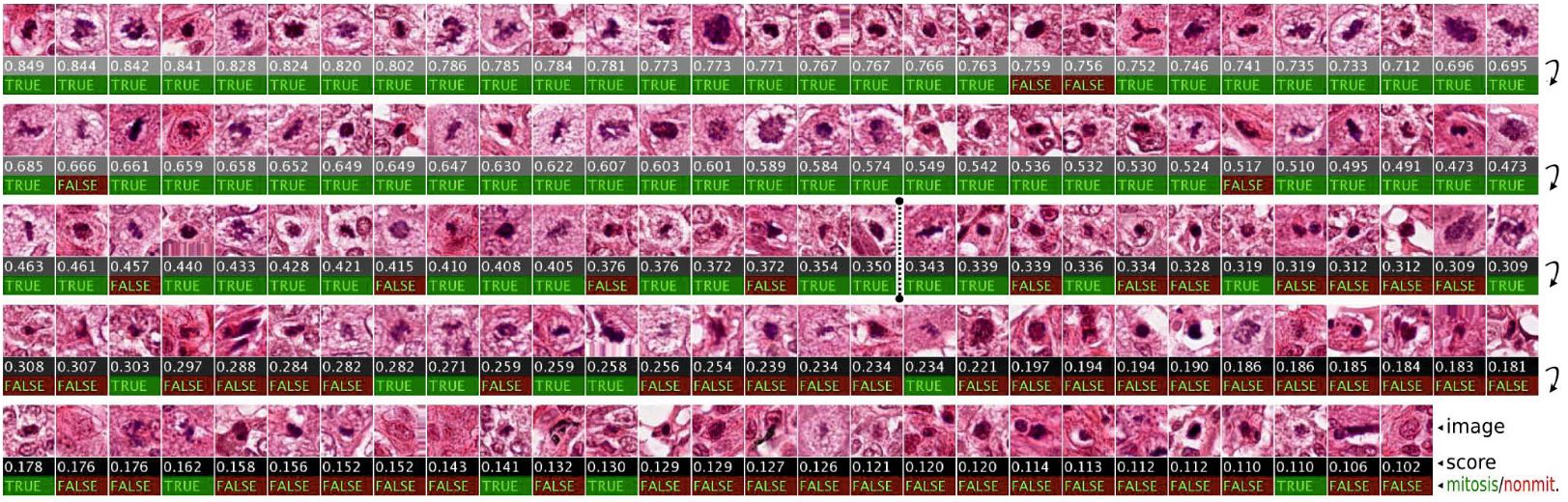
Cireşan's results were far from perfect, but they were impressive enough to be a milestone in the domain: as a winner of the "ICPR 2012 mitosis detection competition", it got a lot of attention... despite the many methodogical issues with the competition itself, which is the topic for another post.
By showing that Deep Learning was a way to get good results on digital pathology tasks, Cireşan and Cruz-Roa opened up the floodgates. Litjens lists many different applications in the subsequent years: bacterial colony counting, classification of mitochondria, classification and grading of different types of cancer, detection of metastases... Mostly on H&E images, but also sometimes using immunohistochemistry, Deep Learning invaded the domain.
A few highly influential works that I would like to mention here, and that I will probably write about more later:
After these pioneering works, the future of the field may seem a little dull. If we have deep neural network that work for most digital pathology tasks... what is there left to do?
Fortunately, finding a good "deep learning network" is only a part of the "digital pathology pipeline". Everything that comes around the network - from the constitution of the datasets to the way the results are evaluated - is often more important to the final result. That is going to be a large part of what I will write about in future posts. Questions surrounding how data from challenges and data from real-world application may differ, questions about the way we evaluate algorithms, about how we declare winners and losers in ways that may not always reflect how useful the algorithms really are. For that, a good starting point will be to take a closer look at the aforementioned MITOS12 challenge from ICPR 2012.